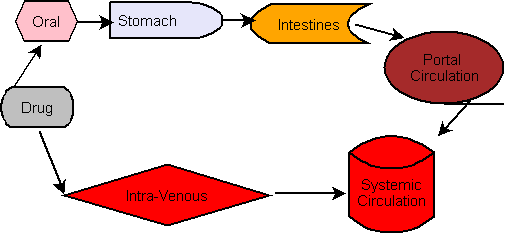
Drug Interactions between Nine Antifungal Agents and Drugs Metabolized by Human Cytochromes P450
Tuesday 7 April 2015
Posted by Unknown
This article reviews in vitro metabolic and in vivo pharmacokinetic drug–drug interactions of nine antifungal agents: six azoles (fluconazole, itraconazole, ketoconazole, miconazole, posaconazole, and voriconazole) and three echinocandins (anidulafungin, caspofungin, and micafungin). In in vitro interaction studies, itraconazole, ketoconazole, and miconazole were found to have higher inhibitory effects on cytochrome P450 (P450 or CYP) 3A4 and 3A5 activities than the other azoles or echinocandins did. Fluconazole, itraconazole, and voriconazole were relatively less potent inhibitors of CYP3A5 than of CYP3A4. The inhibitory effects of fluconazole, itraconazole, ketoconazole, and voriconazole against CYP3A4 and CYP3A5 seemed to be correlated with their dissociation constants for CYP51 (lanosterol 14α-demethylase) from Candida albicans.
In in vivo pharmacokinetic studies, itraconazole was found to be a potent clinically important inhibitor of CYP3A4/5 substrates, and fluconazole and voriconazole increased the blood/plasma concentrations of not only CYP3A4/5 substrates but also CYP2C9 substrates. Miconazole was a potent inhibitor of all P450s investigated in vitro, although there are few detailed studies on the clinical significance of this except for CYP2C9. For the echinocandins, no marked inhibition of P450 activities, except for some inhibition of CYP3A4/5 activity, was observed in vitro. The blood/plasma concentrations of concomitant drug metabolism reviews were not markedly affected by coadministration of echinocandins in vivo, suggesting that echinocandins do not cause clinically significant interactions with drugs that are metabolized by P450s via the inhibition of metabolism. The differential effects of these antifungal agents on P450 activities must be considered when clinicians select antifungal agents for patients also receiving other drugs.
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science
In in vivo pharmacokinetic studies, itraconazole was found to be a potent clinically important inhibitor of CYP3A4/5 substrates, and fluconazole and voriconazole increased the blood/plasma concentrations of not only CYP3A4/5 substrates but also CYP2C9 substrates. Miconazole was a potent inhibitor of all P450s investigated in vitro, although there are few detailed studies on the clinical significance of this except for CYP2C9. For the echinocandins, no marked inhibition of P450 activities, except for some inhibition of CYP3A4/5 activity, was observed in vitro. The blood/plasma concentrations of concomitant drug metabolism reviews were not markedly affected by coadministration of echinocandins in vivo, suggesting that echinocandins do not cause clinically significant interactions with drugs that are metabolized by P450s via the inhibition of metabolism. The differential effects of these antifungal agents on P450 activities must be considered when clinicians select antifungal agents for patients also receiving other drugs.
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science
Epigenetic Regulation of Genes Encoding Drug-Metabolizing Enzymes and Transporters; DNA Methylation and Other Mechanisms
Tuesday 31 March 2015
Posted by Unknown
Drug metabolizing enzymes and transporters are increasingly recognized as key determinants of the inter-individual variability in pharmacokinetic (PK) and pharmacodynamic (PD) outcomes of clinically important drugs. To date, most studies investigating this variability have focused on polymorphisms (e.g. SNPs) in the genes encoding metabolic enzymes and transporters; however, it has recently been reported that the expression of some of these genes is under the control of epigenetic mechanisms.
The most common epigenetic mechanism of mammalian genome regulation is DNA methylation, which does not change the genetic code but affects gene expression. Owing to its maintenance of the genomic sequence, DNA methylation is expected to offer an explanation for the controversial phenotypes of certain genetic polymorphisms. It has been recognized that DNA methylation plays a role in the transcriptional regulation of some PK/PD genes. In this review, we describe the impact of various epigenetic mechanisms, especially DNA methylation, on the expression (or activity) of drug metabolism enzymes and transporter genes.Allogenic hematopoietic stem cell transplantation (HSCT) is a well established but complex treatment option for malignant and non-malignant disorders in pediatric patients.
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science
The most common epigenetic mechanism of mammalian genome regulation is DNA methylation, which does not change the genetic code but affects gene expression. Owing to its maintenance of the genomic sequence, DNA methylation is expected to offer an explanation for the controversial phenotypes of certain genetic polymorphisms. It has been recognized that DNA methylation plays a role in the transcriptional regulation of some PK/PD genes. In this review, we describe the impact of various epigenetic mechanisms, especially DNA methylation, on the expression (or activity) of drug metabolism enzymes and transporter genes.Allogenic hematopoietic stem cell transplantation (HSCT) is a well established but complex treatment option for malignant and non-malignant disorders in pediatric patients.
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science
Targeting Polyamine Metabolism for Finding New Drugs Against Leishmaniasis: A Review
Thursday 26 March 2015
Posted by Unknown
Leishmaniasis is a neglected disease affecting more than 12 million people worldwide. The most used drugs are pentavalent antimonials that are very toxic and display the problem of drug resistance, especially in endemic regions such as Bihar in India. For this reason, it is urgent to find new and less toxic drugs against leishmaniasis. To this end, the understanding of pathways affecting parasite survival is of prime importance for targeted drug discovery. The parasite survival inside the macrophage is strongly dependent on polyamine metabolism. Polyamines are, in fact, very important for cell growth and proliferation. In particular, spermidine (Spd), the final product of the polyamine biosynthesis pathway, serves as a precursor for trypanothione (N1,N8- bis(glutathionyl)spermidine, T(SH)2) and hypusine (Nε-(4-amino-2-hydroxybutyl)lysine). T(SH)2 is a key molecule for parasite defense against the hydrogen peroxide produced by macrophages during the infection. Hypusination is a posttranslational modification occurring exclusively in the eukaryotic initiation factor 5A (eIF5A), which has an important role in avoiding the ribosome stalling during the biosynthesis of protein containing polyprolines sequences. The enzymes, belonging to the spermidine metabolism of drugs, i.e. arginase (ARG), ornithine decarboxylase (ODC), S-adenosylmethionine decarboxylase (AdoMetDC), spermidine synthase (SpdS), trypanothione synthetase (TryS or TSA), trypanothione reductase (TryR or TR), tryparedoxin peroxidase (TXNPx), deoxyhypusine synthase (DHS) and deoxyhypusine hydroxylase (DOHH) are promising targets for the development of new drugs against leishmaniasis. This minireview furnishes a picture of the structural, functional and inhibition studies on polyamine metabolism enzymes that could guide the discovery of new drugs against leishmaniasis.
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science
Research into the genetic basis of schizophrenia is advancing rapidly. This review gives a broad overview of results from successive phases of studies in this field, linking these with recent findings and likely future research directions. Among recent findings, large-scale epidemiological studies based on Scandinavian population registers, have provided further evidence of substantial heritability and evidence that a wide range of psychotic and non-psychotic disorders partly share genetic risk factors with schizophrenia. In molecular genetics, large collaborative genomewide association studies (GWAS) are providing evidence of common risk variants, each of small effect, and many more variants are likely to be found as samples sizes increase further.
A range of rarer Chromosomal disorders copy number variants (CNVs) have been associated with schizophrenia, and both GWAS and CNV studies have provided molecular evidence of genetic overlap between schizophrenia and other disorders. There is increasing interest in phenotypes beyond diagnosis, including further clinical variables and endophenotypes. Next-generation sequencing studies are beginning, with the potential for fast, inexpensive sequencing of the whole genome in large samples, and there is an increasing focus on the functional effects of the candidate risk variants that are being identified.
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science
A range of rarer Chromosomal disorders copy number variants (CNVs) have been associated with schizophrenia, and both GWAS and CNV studies have provided molecular evidence of genetic overlap between schizophrenia and other disorders. There is increasing interest in phenotypes beyond diagnosis, including further clinical variables and endophenotypes. Next-generation sequencing studies are beginning, with the potential for fast, inexpensive sequencing of the whole genome in large samples, and there is an increasing focus on the functional effects of the candidate risk variants that are being identified.
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science
Xenobiotic Metabolism in Human Skin and 3D Human Skin Reconstructs: A Review
Saturday 21 March 2015
Posted by Unknown
In this review, we discuss and compare studies of xenobiotic metabolism in both human skin and 3D human skin reconstructs. In comparison to the liver, the skin is a less studied organ in terms of characterising metabolic capability. While the skin forms the major protective barrier to environmental chemical exposure, it is also a potential target organ for adverse health effects. Occupational, accidental or intended-use exposure to toxic chemicals could result in acute or delayed injury to the skin (e.g. inflammation, allergy, cancer). Skin metabolism may play a role in the manifestation or amelioration of adverse effects via the topical route.
Today, we have robust testing strategies to assess the potential for local skin toxicity of chemical exposure. Such methods (e.g. the local lymph node assay for assessing skin sensitisation; skin painting carcinogenicity studies) incorporate skin metabolism of drugs implicitly in the in vivo model system used. In light of recent European legislation (i.e. 7th Amendment to the Cosmetics Directive and Registration Evaluation and Authorisation of existing Chemicals (REACH)), non-animal approaches will be required to reduce and replace animal experiments for chemical risk assessment. It is expected that new models and approaches will need to account for skin metabolism explicitly, as the mechanisms of adverse effects in the skin are deconvoluted. 3D skin models have been proposed as a tool to use in new in vitro alternative approaches. In order to be able to use 3D skin models in this context, we need to understand their metabolic competency in relation to xenobiotic biotransformation and whether functional activity is representative of that seen in human skin.
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science
Today, we have robust testing strategies to assess the potential for local skin toxicity of chemical exposure. Such methods (e.g. the local lymph node assay for assessing skin sensitisation; skin painting carcinogenicity studies) incorporate skin metabolism of drugs implicitly in the in vivo model system used. In light of recent European legislation (i.e. 7th Amendment to the Cosmetics Directive and Registration Evaluation and Authorisation of existing Chemicals (REACH)), non-animal approaches will be required to reduce and replace animal experiments for chemical risk assessment. It is expected that new models and approaches will need to account for skin metabolism explicitly, as the mechanisms of adverse effects in the skin are deconvoluted. 3D skin models have been proposed as a tool to use in new in vitro alternative approaches. In order to be able to use 3D skin models in this context, we need to understand their metabolic competency in relation to xenobiotic biotransformation and whether functional activity is representative of that seen in human skin.
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science
Inhibitors of the HSP90 Molecular Chaperone: Attacking the Master Regulator in Cancer
Thursday 19 March 2015
Posted by Unknown
The heat shock protein 90 (HSP90) chaperones represent some 1-2% of all cellular protein and are key players in protein quality control in cells. They are over expressed in many human cancers and the fact that many oncogenic proteins are clients has prompted much recent research on HSP90 inhibitors as new cancer therapeutics. A brief introduction is followed by a detailed review of the various classes of inhibitors, both natural product-based and synthetic, that have emerged over the last decade. The natural products geldanamycin, radicicol and novobiocin have provided the start points for new drugs in this area and their medicinal chemistry is reviewed, including the exciting recent results emerging from clinical trials using geldanamycin analogues.
The detailed understanding of the binding mode of these compounds to HSP90 has been significantly enhanced by X-ray crystallography of HSP90 constructs co-crystallised with various ligands. Efforts to replace the natural product inhibitors with more drug-like synthetic compounds have mushroomed over the last 4 years. The purines and the 3,4-diarylpyrazoles have proven to be the most successful and their Medicinal Chemistry is reviewed with particular emphasis on structure-based design. Protein/ligand co-crystal structures have shown that conserved water molecules in the active site are a vital part of the hydrogen-bonding network established on binding both natural product and synthetic inhibitors. Medicinal chemists have used this information to develop high affinity lead compounds. Recent research provides the platform for exciting developments in the area of HSP90 inhibition over the next few years.
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science
The detailed understanding of the binding mode of these compounds to HSP90 has been significantly enhanced by X-ray crystallography of HSP90 constructs co-crystallised with various ligands. Efforts to replace the natural product inhibitors with more drug-like synthetic compounds have mushroomed over the last 4 years. The purines and the 3,4-diarylpyrazoles have proven to be the most successful and their Medicinal Chemistry is reviewed with particular emphasis on structure-based design. Protein/ligand co-crystal structures have shown that conserved water molecules in the active site are a vital part of the hydrogen-bonding network established on binding both natural product and synthetic inhibitors. Medicinal chemists have used this information to develop high affinity lead compounds. Recent research provides the platform for exciting developments in the area of HSP90 inhibition over the next few years.
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science
On the Structural Plasticity of the Human Genome: Chromosomal Inversions Revisited
Wednesday 18 March 2015
Posted by Unknown
With the aid of novel and powerful molecular biology techniques, recent years have witnessed a dramatic increase in the number of studies reporting the involvement of complex structural variants in several genomic disorders. In fact, with the discovery of Copy Number Variants (CNVs) and other forms of unbalanced structural variation, much attention has been directed to the detection and characterization of such rearrangements, as well as the identification of the mechanisms involved in their formation. However, it has long been appreciated that chromosomes can undergo other forms of structural changes - balanced rearrangements - that do not involve quantitative variation of genetic material.
Indeed, a particular subtype of balanced rearrangement – inversions – was recently found to be far more common than had been predicted from traditional cytogenetics. Chromosomal disorders inversions alter the orientation of a specific genomic sequence and, unless involving breaks in coding or regulatory regions (and, disregarding complex trans effects, in their close vicinity), appear to be phenotypically silent. Such a surprising finding, which is difficult to reconcile with the classical interpretation of inversions as a mechanism causing subfertility (and ultimately reproductive isolation), motivated a new series of theoretical and empirical studies dedicated to understand their role in human genome evolution and to explore their possible association to complex genetic disorders. With this review, we attempt to describe the latest methodological improvements to inversions detection at a genome wide level, while exploring some of the possible implications of inversion rearrangements on the evolution of the human genome.
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science
Indeed, a particular subtype of balanced rearrangement – inversions – was recently found to be far more common than had been predicted from traditional cytogenetics. Chromosomal disorders inversions alter the orientation of a specific genomic sequence and, unless involving breaks in coding or regulatory regions (and, disregarding complex trans effects, in their close vicinity), appear to be phenotypically silent. Such a surprising finding, which is difficult to reconcile with the classical interpretation of inversions as a mechanism causing subfertility (and ultimately reproductive isolation), motivated a new series of theoretical and empirical studies dedicated to understand their role in human genome evolution and to explore their possible association to complex genetic disorders. With this review, we attempt to describe the latest methodological improvements to inversions detection at a genome wide level, while exploring some of the possible implications of inversion rearrangements on the evolution of the human genome.
For a complete list, click on Bentham Science Publishers’ Journals Impacting Science